- ERAD koneistosta artikkeleita.
- ERAD tarkoittaa endoplasmiseen retikulumiin(ER) assosioitunutta hajoitusjärjestelmää, joka on solutalouden kannalta eräänlainen tarkka hyödynnyslaitos tekovikamaterialle, jota ei onnistuta korjaamaan.
- Virukset ja toksiinit kaappaavat tätä järjestelmää.
https://www.sciencedirect.com/science/article/pii/S1097276515004499
ER-associated
degradation (ERAD) is a protein clearance mechanism by which misfolded,
misassembled, or metabolically regulated proteins are specifically
dislocated from the ER into the cytosol and degraded by the ubiquitin proteasome system.
However, the ironic truth is that membrane-penetrating transportation and protein degradation
machineries in ERAD are preferably hijacked by exogenous pathogens such
as viruses and toxins for their invasion and evasion from immunological surveillance.
In this Review, we provide an overview of our current understanding of
the pathogenic hijacking of the host cell ERAD, in which pathogens
exploit the complex ERAD machinery in a variety of manners for their own
use, suggesting flexibility and plasticity of the molecular machinery
of ERAD.
- ERAD koneisto toimii endoplasmisen retikulumin (ER) syntetisoimien proteiinin laadunkontrollijärjestelmänä (QC) (Quality Control )
- Tämä ERQC käsittää useita vaiheita ja lukuisia komponentteja.
ERAD functions in ER protein quality control (ERQC) and sterol regulation (Ruggiano et al., 2014).
Over the past two decades, it has been elucidated that ERQC-related
ERAD is a multi-step and branched process that has numerous components (Araki and Nagata, 2011, Christianson and Ye, 2014).
ERQC kolme akselia.
(Chaperoneproteiini = kaitsijaproteiini).
I chaperone-proteiiniryhmä on immunoglobuliinin raskaan ketjun sisältävä BiP, joka kuuluu HSP70-proteiiniperheeseen ja se edistää proteiinin laskostumista sisäisellä ATP.ta hydroplysoivalla aktiivisuudella.
II chaperoneproteiiniryhmä ovat ER oxidoreduktaasit, kuten
PDI, proteiinidisulfidi-isomeraasi;
ERO1, ER oxidoreductiini-1.
ja ERp57, joka oksidoi cysteiinitähteitä , siis muodostaa niissä disulfidisiltoja (-S-S-).
III chaperoniproteiiniryhmä ovat kalnexiini (CNX) ja kalretikuliini(CRT). Ne ovat lektiinien kaltaisia kaitsijoita ja ne tunnistavat oligosakkaridiketjuja vastasyntetisoituneissa glykoproteiineissa ja edistävät proteiinin laskostumista.
Eräät näistä mainituista tekijöistä osallistuvat myös ERAD.iin ja moni patogeeni hyödyntää niitä. ( Esim SARSc tekee interaktion ER- kalvoon integroituneeseen kalnexiiniin , mutta välttää interaktiota luminaaliseen kalretikuliiniin, jolloin se häiritsee CRT/CNX toimintasykliä).
https://kyushu-u.pure.elsevier.com/en/publications/monitoring-of-s-protein-maturation-in-the-endoplasmic-reticulum-b
ERQC kolme akselia.
Three Axes of ERQC
I akseli on proteiinien laskostuminen. Useimmat Endoplasmisessa retikulumissa (ER) syntetisoituneista proteiineista ovat modifioitu kovalenttisesti molekyylien nsisäisten ja välisten disulfidisiltojen (-S-S-) ja oligosakkaridiketjujen avulla (OS) samanaikaisesti kun peptidiä syntetuisoidaan ja laskostetaan. Näitten modifikaatioitten avulla proteiinit saavat sellaista kestokykyä, jolla ne pysyvät elossa myös solun ulkopuolisessa miljöössä. Tähän modifikaatioon kytkeytyneeseein proteiinien laskostumiseen osallistuu kolme molekulaarista kaitsijaproteiiniryhmää(Chaperoneproteiini = kaitsijaproteiini).
I chaperone-proteiiniryhmä on immunoglobuliinin raskaan ketjun sisältävä BiP, joka kuuluu HSP70-proteiiniperheeseen ja se edistää proteiinin laskostumista sisäisellä ATP.ta hydroplysoivalla aktiivisuudella.
II chaperoneproteiiniryhmä ovat ER oxidoreduktaasit, kuten
PDI, proteiinidisulfidi-isomeraasi;
ERO1, ER oxidoreductiini-1.
ja ERp57, joka oksidoi cysteiinitähteitä , siis muodostaa niissä disulfidisiltoja (-S-S-).
III chaperoniproteiiniryhmä ovat kalnexiini (CNX) ja kalretikuliini(CRT). Ne ovat lektiinien kaltaisia kaitsijoita ja ne tunnistavat oligosakkaridiketjuja vastasyntetisoituneissa glykoproteiineissa ja edistävät proteiinin laskostumista.
Eräät näistä mainituista tekijöistä osallistuvat myös ERAD.iin ja moni patogeeni hyödyntää niitä. ( Esim SARSc tekee interaktion ER- kalvoon integroituneeseen kalnexiiniin , mutta välttää interaktiota luminaaliseen kalretikuliiniin, jolloin se häiritsee CRT/CNX toimintasykliä).
https://kyushu-u.pure.elsevier.com/en/publications/monitoring-of-s-protein-maturation-in-the-endoplasmic-reticulum-b
- Monitoring of s protein maturation in the endoplasmic reticulum by calnexin is important for the infectivity of severe acute respiratory syndrome coronavirus Masaya Fukushi, Yoshiyuki Yoshinaka et al Severe acute respiratory syndrome coronavirus (SARS-CoV) is the etiological agent of SARS, a fatal pulmonary disorder with no effective treatment. We found that SARS-CoV spike glycoprotein (S protein), a key molecule for viral entry, binds to calnexin, a molecular chaperone in the endoplasmic reticulum (ER), but not to calreticulin, a homolog of calnexin. . These findings demonstrated that calnexin strictly monitors the maturation of S protein by its direct binding, resulting in conferring infectivity on SARS-CoV.
The first axis of ERQC is protein folding (Araki and Nagata, 2011). Most newly synthesized proteins in the ER are covalently modified with intra- and/or inter-molecular disulfide bonds and oligosaccharide
chains in parallel with their synthesis and folding. These
modifications reinforce the structure of these proteins such that they
can survive in the extracellular environment. Three groups of molecular
chaperones and folding enzymes majorly contribute to
modification-coupled protein folding. The first group includes classic
molecular chaperones, such as immunoglobulin heavy chain-binding protein
(BiP), which belongs to the heat shock protein 70 family and promotes protein folding with an intrinsic ATP-hydrolyzing activity (Otero et al., 2010). The second group consists of ER oxidoreductases, such as protein disulfide isomerase (PDI), ER oxidoreductin 1 (ERO1), and ERp57, which oxidize substrate cysteine
residues (synonymous with the formation of a disulfide bond) and
isomerize improperly oriented disulfide bonds in parallel with de novo
protein synthesis and folding (Ellgaard and Ruddock, 2005). The third group consists of lectin-like chaperones such as calnexin and calreticulin, which recognize oligosaccharide chains attached to newly synthesized glycoproteins and promote protein folding (Helenius and Aebi, 2004).
Some of these factors also participate in ERAD and are occasionally utilized by pathogens, as discussed later.
II akseli on vaste proteiinin laskostumattomuuteen. (UPR, Unfolded protein response).
Jos jostain syystä proteiinit eivät laskostu tai laskostuvat väärin ja jos tällaista materiaalia kertyy endoplasmiseen verkostoon ylen määrin, ylittyy kapasiteetti saada tätä materiaalia pois endoplasmisesta verkostosta.
Näitä proteiineja, joiden tehtäviin kuluu tunnistaa laskostumattomien proteiinien ylikuormitusta, ovat PERK, ATF6 ja IRE1, ja ne pystyvät kukin tiettyä tietään aloittamaan alavirran tapahtumia kuten translaation vaimentamista ja laskostavien entsyymien ERAD- tekijöiden ylössäätämistä jotta ER ylikuormitus tila korjautuisi ja ER proteostaasi palautuisi. ESIM: https://www.frontiersin.org/articles/10.3389/fendo.2018.00210/full
https://www.frontiersin.org/files/Articles/364990/fendo-09-00210-HTML/image_m/fendo-09-00210-g001.jpg
Näitä proteiineja, joiden tehtäviin kuluu tunnistaa laskostumattomien proteiinien ylikuormitusta, ovat PERK, ATF6 ja IRE1, ja ne pystyvät kukin tiettyä tietään aloittamaan alavirran tapahtumia kuten translaation vaimentamista ja laskostavien entsyymien ERAD- tekijöiden ylössäätämistä jotta ER ylikuormitus tila korjautuisi ja ER proteostaasi palautuisi. ESIM: https://www.frontiersin.org/articles/10.3389/fendo.2018.00210/full
https://www.frontiersin.org/files/Articles/364990/fendo-09-00210-HTML/image_m/fendo-09-00210-g001.jpg
The second axis of ERQC is the unfolded protein response
(UPR). When the level of unfolded or misfolded proteins accumulated in
the ER exceeds the folding and clearance capacity of the ER for any
reason, particular ER membrane proteins such as PERK, ATF6, and IRE1 sense the overloaded state and initiate downstream events, including translation attenuation and upregulation of folding enzymes and ERAD factors, which comprehensively reduce the protein burden in the ER and restore ER proteostasis (see previous review articles, e.g., Walter and Ron, 2011). The UPR also involves an apoptotic pathway, which is activated when the cell fails to resolve the stress conditions (Walter and Ron, 2011).
III akseli on varsinainen ERAD.
Finally, the third axis of ERQC is ERAD, which is described in the next section.
(Tähän laitan ERAD kohtaan kuuluvia SARS interaktioproteiineja sitä mukaa kuin löydän. Selenos eli VIMP on yksi näitä.
https://www.researchgate.net/publication/307442236/figure/fig1/AS:403388127825920@1473186632494/ER-associated-degradation-ERAD-pathway-ERAD-is-an-ER-quality-control-pathway-that.png
SELENOS (15q26.2 Aliases for SELENOS Gene, VIMP
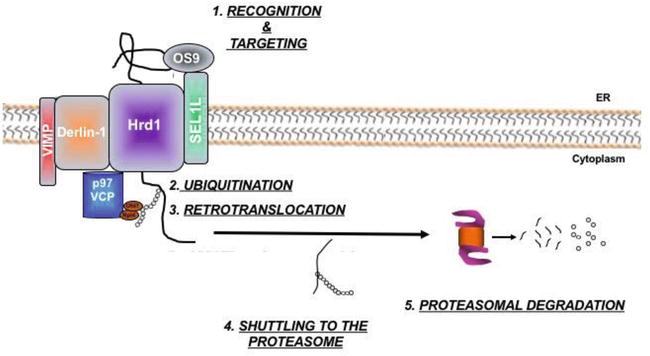
This gene SELENOS encodes a transmembrane protein that is localized in the endoplasmic reticulum (ER). It is involved in ERAD, the degradation process of misfolded proteins in the ER, and may also have a role in inflammation control. This protein is a selenoprotein, containing the rare amino acid selenocysteine (Sec). Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. Two additional phylogenetically conserved stem-loop structures (Stem-loop 1 and Stem-loop 2) in the 3' UTR of this mRNA have been shown to function as modulators of Sec insertion. An alternatively spliced transcript variant, lacking the SECIS element and encoding a non-Sec containing shorter isoform, has been described for this gene (PMID:23614019). [provided by RefSeq, Jul 2017]
https://www.researchgate.net/publication/307442236/figure/fig1/AS:403388127825920@1473186632494/ER-associated-degradation-ERAD-pathway-ERAD-is-an-ER-quality-control-pathway-that.png
SELENOS (15q26.2 Aliases for SELENOS Gene, VIMP
Selenoprotein S
2
3
4
5
VCP-Interacting Membrane Protein 2 3 4 Valosin-Containing Protein-Interacting Membrane Protein 2 3 VCP Interacting Membrane Selenoprotein 2 3
VIMP 3 4, AD-015 3, ADO15 3, SBBI8 3 SEPS1 3, Tanis 3 , SelS 4, SELS 3 4
VCP-Interacting Membrane Protein 2 3 4 Valosin-Containing Protein-Interacting Membrane Protein 2 3 VCP Interacting Membrane Selenoprotein 2 3
VIMP 3 4, AD-015 3, ADO15 3, SBBI8 3 SEPS1 3, Tanis 3 , SelS 4, SELS 3 4

This gene SELENOS encodes a transmembrane protein that is localized in the endoplasmic reticulum (ER). It is involved in ERAD, the degradation process of misfolded proteins in the ER, and may also have a role in inflammation control. This protein is a selenoprotein, containing the rare amino acid selenocysteine (Sec). Sec is encoded by the UGA codon, which normally signals translation termination. The 3' UTRs of selenoprotein mRNAs contain a conserved stem-loop structure, designated the Sec insertion sequence (SECIS) element, that is necessary for the recognition of UGA as a Sec codon, rather than as a stop signal. Two additional phylogenetically conserved stem-loop structures (Stem-loop 1 and Stem-loop 2) in the 3' UTR of this mRNA have been shown to function as modulators of Sec insertion. An alternatively spliced transcript variant, lacking the SECIS element and encoding a non-Sec containing shorter isoform, has been described for this gene (PMID:23614019). [provided by RefSeq, Jul 2017]
Inga kommentarer:
Skicka en kommentar