https://go.gale.com/ps/i.do?p=AONE&u=googlescholar&id=GALE|A195522200&v=2.1&it=r&sid=AONE&asid=9f6766c0
Main content
From December 2003 through January 2004, the Phnom Tamao Wildlife
Rescue Centre, Cambodia, was affected by the highly pathogenic influenza
virus (H5N1). Birds from 26 species died. Influenza virus subtype H5N1 was
detected in 6 of 7 species tested. Cats from 5 of 7 species were probably
infected; none died.
On January 24, 2004, the first confirmed outbreak of highly
pathogenic avian influenza virus (HPAIV) subtype H5N1 in Cambodia was
reported to the Office International des Epizooties (1). During the previous
month, an unusually high mortality rate had been noted among captive wild
birds at the Phnom Tamao Wildlife Rescue Centre (PTWRC) in Takeo Province, 45
km South from Phnom Penh. We report the results of a retrospective
investigation of this outbreak.
During the outbreak period, PTWRC housed 600-1,000 wild animals
(70 species of mammals, birds, and reptiles). The center is divided into 3
main sections that cover 37 ha. Birds were kept in sections S1-1, S1-2, and
S2, and the cats were in all sections (Figure). The information on bird
deaths at PTWRC was systematically recorded by WildAid staff members who were
at the Centre at the time of the outbreak. In June 2004, a complete
investigation was conducted at PTWRC, and semistructured interviews of key
informants were used to identify deaths of domestic poultry in the
surrounding villages. Every bird death between December 15, 2003, through
January 15, 2004, was defined as a suspected case of HPAIV (H5N1). For S1,
the cumulative mortality rate could not be estimated because the exact bird
population was not known and the birds were difficult to observe in that
section (the semicaptive waterfowl population is able to mix with the wild
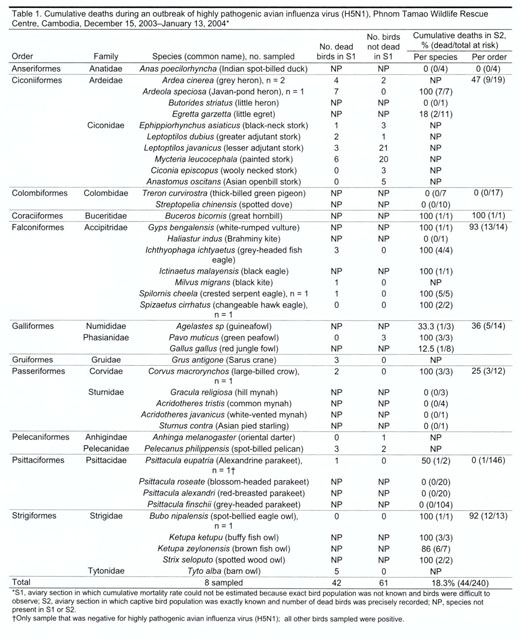
population and disperse to breed). For S2, information was complete (Table
1).
The first case, in a crested serpent eagle (Spilornis cheela), was
reported on December 15, 2003, in S2 (Figure). On December 19, the outbreak
had reached every section and continued until January 12; a total of 86
birds, representing 8 taxonomic orders and 12 families, died (Table 1). Of 7
cat species, cats from 5 species were reported sick (16/39 total cats) (Table
2). In $2, 80% of the reported bird deaths were observed from December 15 to
21. Of the 29 wild bird species kept in $2 at the beginning of the outbreak,
no birds from 12 species showed signs of disease (Table 1). Mortality rates
varied among the orders, 0-100% (Table 1). The only mammals present in the
aviaries in $2, slow lorises (Nycticebus sp.), did not become ill. None of
the 27 animal keepers, who were 20-50 years of age, were reported to have
gotten sick.
Most of the birds died within a few hours without showing any
clinical signs of infection. A few birds died 1-2 days after onset of
clinical signs (anorexia, extreme lethargy, occasional dark green diarrhea,
respiratory distress, and neurologic abnormalities). The cats were sick for
5-7 days and exhibited anorexia and lethargy but no respiratory illness.
Laboratory investigations of the organs from 8 birds sampled in
December 2003 were performed (Table 1). For those birds, West Nile virus
infection was ruled out by reverse transcription-PCR (RT-PCR), according to
the procedure described by Lanciotti et al. (2). All birds sampled, except a
parakeet, were positive for influenza subtype H5N1 by RT-PCR (3) (Table 1).
Molecular characterizations of hemagglutinin (H)5 and neuraminidase (N)1 were
performed from the influenza virus (H5N1) strains from PTWRC as previously
described (4). H5 amino acid sequences were identical in the coding region to
the sequence of isolates obtained from poultry cases in Cambodia (ill poultry
from a flock with high mortality rates) and similar ([greater than or equal
to]96.5%) to HPAIV (H5N1) strain H5 sequences from Vietnam and Thailand in
2004 (data not shown). All belonged to the H5 clade 1 (4). Amino acid
sequences from N 1 from Cambodia were very close to each other ([greater than
or equal to]97.12% identity) and to 2004 Vietnamese and Thai N1 sequences
(>96%) (data not shown). The HA and NA sequences of the isolates were
deposited in GenBank (accession nos. ISDN186319-ISN186324, ISDN186329,
ISDNI86330ISDN 186665, and ISDN242365).
Retrospective investigation of the villages surrounding the PTWRC
and Phnom Perth showed that chickens from 2 flocks in which deaths had been
reported in mid-December had been provided to the PTWRC, either for the
restaurants or for the captive animal feeding. Furthermore, at the time of
the outbreak, many wild crows were found dead in the forest surrounding the
PTWRC.
The 4 cat serum samples, each from a different species, were
positive for HPAIV (H5N1) with serum neutralization test (5); titers ranged
from 10 to 40 (Table 2). None of the affected cats died.
The sources of introduction of HPAIV (H5N1) within the PTWRC were
probably multiple: virus-infected chicken bought to feed the carnivorous
species, infected live chickens brought to restaurants near S2 (i.e., the
first place where deaths were detected), and contact between infected wild
and captive birds. The introduction through infected chickens is supported by
the absence of an outbreak at the PTWRC after the feeding of chickens to
carnivorous species was discontinued; however, deaths in domestic poultry
continued in the area. In addition, almost all carnivorous bird species in S2
died (93% of Falconiformes and 92% of Strigiformes) as did most species
usually fed chicken meat in captivity (herons, storks, crows, great hornbill,
pelican). Diet was also the origin of the outbreak among tigers and leopards
in Thailand (6,7). The dispersion of the disease between PTWRD sections was
probably due to poor biosecurity measures.
The clinical outcome of wild birds with suspected HPAIV (H5N1)
infection at PTWRC ranged from severe illness and death to complete absence
of clinical signs, as described (8). Several species from the orders
Ciconiiformes, Galliformes, Passeriformes, Gruiformes, Coraciiformes, and
Pelecaniformes were affected during the outbreak. This observation is
consistent with data published earlier, except for Coraciiformes represented
by 1 bird in our study (9). Only the carnivorous species (Corvus
macrorynchos) among the 5 species of Passeriformes in the aviaries showed
clinical signs and later was confirmed by RT-PCR to be positive for HPAIV
(H5N1). This outbreak confirms that Falconiformes and Strigiformes are
sensitive to HPAIV (H5N1) infection and disease (10-12) and shows that
numerous species of these orders can be affected by HPAIV (H5N1) (Table 1).
Psittaciformes and Columbiformes were not visibly affected by the outbreak
although they were kept in large numbers in S2, where large numbers of deaths
occurred. As non-water-bird species, they do not belong to groups in which
avian influenza is commonly reported (13). Anseriformes, represented in PTWRC
by only 4 birds (Anas poecilorhyncha), did not show any clinical signs.
Heterogeneity in the susceptibility of wild ducks to HPAIV (H5N1), including
asymptomatic infection, has been demonstrated (14); this species also belongs
to the group of wild ducks found asymptomatically infected with HPAIV (H5N1)
in the People's Republic of China during the winter of 2005 (15).
The serologic evidence of influenza virus (H5N1) infection in 4
species of wild cats is in agreement with previous infection in Thailand (6,
7). The report of illness in the Asiatic golden cat (Catopuma temminckii) and
the clouded leopard (Neofelis nebulosa) broadens the host range of the virus
among mammals.
This report confirms the great variability of wild bird and mammal
responses to HPAIV (H5N1) infection. It also confirms the broadening range of
susceptible species that may be specific to this clade 1 virus.
References
(1.) World Organisation for Animal Health. Update on highly
pathogenic avian influenza in animals (type H5 and H7) [cited 2008 May 31].
Available from http://www.oie.int/downld/AVIAN%20INFLUENZA/ A2004_AI.php
(2.) Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vance-Vorndam
A. Rapid detection and typing of dengue viruses from clinical samples by
using reverse transcriptase-polymerase chain reaction. J Clin Microbiol.
1992;30:545-51.
(3.) World Organisation for Animal Health. Recommended laboratory
tests to identify influenza A/H5 virus in specimens from patients with an
influenza-like illness. 19 February 2004 [cited 2005 Jun 12]. Available from
http://www.who.int/csr/disease/avian_influenza/
guidelines/en/avian_labtests1.pdf
(4.) The World Health Organization Global Influenza Program
Surveillance Network. Evolution of H5N1 avian influenza viruses in Asia.
Emerg Infect Dis. 2005;11:1515-21.
(5.) Choi YK, Nguyen TD, Ozaki H, Webby RJ, Puthavathana P,
Buranathal C, et al. Studies of H5NI influenza virus infection of pigs by
using viruses isolated in Vietnam and Thailand in 2004. J Virol.
2005;79:10821-5. DOI: 10.1128/JV1.79.16.10821-10825.2005
(6.) Keawcharoen J, Oraveerakul K, Kuiken T, Fouchier RAM, Amonsin
A, Payungporn S, et al. Avian influenza H5N1 in tigers and leopards. Emerg
Infect Dis. 2004;10:2189-9l. DOI: 10.1007/b100517
(7.) Thanawongnuwech R, Amonsin A, Tantilertcharoen R,
Damrongwatanapokin S, Theamboonlers A, Payungporn S, et al. Probable
tiger-to-tiger transmission of avian influenza H5N1. Emerg Infect Dis.
2005;11:699-701. DOI: 10.1007/b102143
8.) Webster RG, Hulse DJ. Microbial adaptation and change: avian
influenza. Rev Sci Tech. 2004;23:453-65. DOI: 10.2172/15009826
(9.) Whitworth D, Newman SH, Mundkur T, Harris P, eds. Wild birds
and avian influenza: an introduction to applied field research and disease
sampling techniques. Rome (Italy): Food and Agriculture Organization, Animal
Production and Health; 2007 [cited 2008 Jun 2]. Available from
http://www.fao.org/avianflu/en/wildlife/info_res.htm
(10.) World Organisation for Animal Health. Avian influenza in
Hong Kong (Special Administrative Region of the People's Republic of
China) in a wild bird [in French]. Informations Sanitaires. 2004;17 [cited
2009 Jan 21]. Available from
ftp://ftp.oie.int/infos_san_archives/fr/2004/fr_040130v17n05.pdf
(11.) Van Bonn S, Thomas I, Hanquet G, Lambrecht B, Boschmans M,
Dupont G, et al. Highly pathogenic H5N1 influenza virus in smuggled Thai
eagles, Belgium. Emerg Infect Dis. 2005;11:702-5. DOI: 10.1007/b102143
(12.) Sabirovic M, Wilesmith J, Hall S, Coulson N, Landeg F.
Outbreaks of HPAI H5N1 virus in Europe during 2005/2006. DEFRA, 2006 [cited
2009 Jan 21]. Available from http://collections.europarchive.org/
tna/20080107205404/http://defra.gov.uk/
animalh/diseases/monitoring/riskassess.htm
(13.) Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus
AD, Fouchier RA. Global patterns of influenza A virus in wild birds. Science.
2006;312:384-8. DOI: 10.1126/science. 1122438
(14.) Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE.
Susceptibility of North American ducks and gulls to H5N1 highly pathogenic
avian influenza viruses. Emerg Infect Dis. 2006; 12:1663-70.
(15.) Chen H, Smith GJ, Li KS, Wang J, Fan XH, Rayner JM, et al.
Establishment of multiple sublineages of H5NI influenza virus in Asia:
implications for pandemic control. Proc Natl Acad Sci U S A. 2006;103:2845
50. DOI: 10.1073/pnas.0511120103
Address for correspondence: Stephanie Desvaux, CIRAD, AGIRs,
National Institute of Veterinary Research, 86 Truong Chinh, Hanoi, Vietnam;
email: stephanie.desvaux@cirad.fr
Author affiliations: Centre de Cooperation Internationale en
Recherche Agronomique pour le Developpement, Montpellier, France (S. Desvaux,
N. Gaidet); WildAid, Phnom Penh, Cambodia (N. Marx, M. Hunt); Institut
Pasteur du Cambodge, Phnom Penh (S. Ong, J.-M. Reynes); Institut Pasteur,
Paris, France (J.-C. Manuguerra, S. Van der Werf); National Animal Health and
Production Investigation Center, Phnom Penh (S. Sorn); and University of Hong
Kong and Queen Marie Hospital, Pokfulam, Hong Kong Special Administrative
Region, People's Republic of China (M. Peiris)
DOI: 10.3201/eid1503.081410
Dr Desvaux is a veterinary epidemiologist working at the Centre de
Cooperation Internationale en Recherche Agronomique pour le Developpement.
Her current research interests focus on HPAIV epidemiology and surveillance
in Vietnam.
Table 1. Cumulative deaths during an outbreak of highly pathogenic
avian influenza virus (H5N1), Phnom Tamao Wildlife Rescue
Centre, Cambodia, December 15, 2003-January 13, 2004 *
Order Family Species (common name),
no. sampled
Anseriformes Anatidae Anas poecilorhyncha
(Indian spot-billed
duck)
Ciconiiformes Ardeidae Ardea cinerea
(grey heron),
n = 2
Ardeola speciosa
(Javan-pond
heron), n = 1
Butorides striatus
(little heron)
Egretta garzetta
(little egret)
Ciconidae Ephippiorhynchus
asiaticus
(black-neck stork)
Leptoptilos dubius
(greater adjutant
stork)
Leptoptilos javanicus
(lesser adjutant stork)
Mycteria leucocephala
(painted stork)
Ciconia episcopus
(wooly necked stork)
Anastomus oscitans
(Asian openbill
stork)
Colombiformes Colombidae Treron curvirostra
(thick-billed
green pigeon)
Streptopelia chinensis
(spotted dove)
Coraciiformes Buceritidae Buceros bicomis
(great hornbill)
Falconiformes Accipitridae Gyps bengalensis
(white-rumped
vulture)
Haliastur indus
(Brahminy kite)
Ichthyophaga ichtyaetus
(grey-headed fish
eagle)
Ictinaetus malayensis
(black eagle)
Milvus migrans
(black kite)
Spilomis cheela
(crested serpent
eagle), n = 1
Spizaetus cirrhatus
(changeable hawk eagle),
n = 1
Galliformes Numididae Agelastes sp
(guineafowl)
Phasianidae Pavo muticus
(green peafowl)
Gallus gallus
(red jungle fowl)
Gruiformes Gruidae Grus antigone
(Sarus crane)
Passeriformes Corvidae Corvus macrorynchos
(large-billed crow),
n = 1
Sturnidae Gracula religiosa
(hill mynah)
Acridotheres tristis
(common mynah)
Acridotheres javanicus
(white-vented mynah)
Stumus contra (Asian
pied starling)
Pelecaniformes Anhigindae Anhinga melanogaster
(oriental darter)
Pelecanidae Pelecanus philippensis
(spot-billed pelican)
Psittaciformes Psittacidae Psittacula eupatria
(Alexandrine parakeet),
n = 1 ([dagger])
Psittacula roseate
(blossom-headed
parakeet)
Psittacula alexandri
(red-breasted parakeet)
Psittacula finschii
(grey-headed parakeet)
Strigiformes Strigidae Bubo nipalensis
(spot-bellied
eagle owl),
n = 1
Ketupa ketupu
(buffy fish owl)
Ketupa zeylonensis
(brown fish owl)
Strix seloputo
(spotted wood owl)
Tytonidae Tyto alba (barn owl)
Total
8 sampled
No. birds
No. dead not dead
Species (common name), birds in S1 in S1
no. sampled
Anas poecilorhyncha NP NP
(Indian spot-billed
duck)
Ardea cinerea 4 2
(grey heron),
n = 2
Ardeola speciosa 7 0
(Javan-pond
heron), n = 1
Butorides striatus NP NP
(little heron)
Egretta garzetta NP NP
(little egret)
Ephippiorhynchus 1 3
asiaticus
(black-neck stork)
Leptoptilos dubius 2 1
(greater adjutant
stork)
Leptoptilos javanicus 3 21
(lesser adjutant stork)
Mycteria leucocephala 6 20
(painted stork)
Ciconia episcopus 0 3
(wooly necked stork)
Anastomus oscitans 0 5
(Asian openbill
stork)
Treron curvirostra NP NP
(thick-billed
green pigeon)
Streptopelia chinensis NP NP
(spotted dove)
Buceros bicomis NP NP
(great hornbill)
Gyps bengalensis NP NP
(white-rumped
vulture)
Haliastur indus NP NP
(Brahminy kite)
Ichthyophaga ichtyaetus 3 0
(grey-headed fish
eagle)
Ictinaetus malayensis NP NP
(black eagle)
Milvus migrans 1 0
(black kite)
Spilomis cheela 1 0
(crested serpent
eagle), n = 1
Spizaetus cirrhatus 0 0
(changeable hawk eagle),
n = 1
Agelastes sp NP NP
(guineafowl)
Pavo muticus 0 3
(green peafowl)
Gallus gallus NP NP
(red jungle fowl)
Grus antigone 3 0
(Sarus crane)
Corvus macrorynchos 2 0
(large-billed crow),
n=1
Gracula religiosa NP NP
(hill mynah)
Acridotheres tristis NP NP
(common mynah)
Acridotheres javanicus NP NP
(white-vented mynah)
Stumus contra (Asian NP NP
pied starling)
Anhinga melanogaster 0 1
(oriental darter)
Pelecanus philippensis 3 2
(spot-billed pelican)
Psittacula eupatria 1 0
(Alexandrine parakeet),
n = 1 ([dagger])
Psittacula roseate NP NP
(blossom-headed
parakeet)
Psittacula alexandri NP NP
(red-breasted parakeet)
Psittacula finschii NP NP
(grey-headed parakeet)
Bubo nipalensis 0 0
(spot-bellied
eagle owl),
n = 1
Ketupa ketupu NP NP
(buffy fish owl)
Ketupa zeylonensis NP NP
(brown fish owl)
Strix seloputo NP NP
(spotted wood owl)
Tyto alba (barn owl) 5 0
8 sampled 42 61
Cumulative deaths in S2,
% (dead/total at risk)
Species (common name), Per species Per order
no. sampled
Anas poecilorhyncha 0 (0/4) 0 (0/4)
(Indian spot-billed
duck)
Ardea cinerea NP 47 (9/19)
(grey heron),
n = 2
Ardeola speciosa 100 (7/7)
(Javan-pond
heron), n = 1
Butorides striatus 0 (0/1)
(little heron)
Egretta garzetta 18 (2/11)
(little egret)
Ephippiorhynchus NP
asiaticus
(black-neck stork)
Leptoptilos dubius NP
(greater adjutant
stork)
Leptoptilos javanicus NP
(lesser adjutant stork)
Mycteria leucocephala NP
(painted stork)
Ciconia episcopus NP
(wooly necked stork)
Anastomus oscitans NP
(Asian openbill
stork)
Treron curvirostra 0 (0/7 0 (0/17)
(thick-billed
green pigeon)
Streptopelia chinensis 0 (0/10)
(spotted dove)
Buceros bicomis 100 (1/1) 100 (1/1)
(great hornbill)
Gyps bengalensis 100 (1/1) 93 (13/14)
(white-rumped
vulture)
Haliastur indus 0 (0/1)
(Brahminy kite)
Ichthyophaga ichtyaetus 100 (4/4)
(grey-headed fish
eagle)
Ictinaetus malayensis 100 (1/1)
(black eagle)
Milvus migrans NP
(black kite)
Spilomis cheela 100 (5/5)
(crested serpent
eagle), n = 1
Spizaetus cirrhatus 100 (2/2)
(changeable hawk eagle),
n=1
Agelastes sp 33.3 (1/3) 36 (5/14)
(guineafowl)
Pavo muticus 100 (3/3)
(green peafowl)
Gallus gallus 12.5 (1/8)
(red jungle fowl)
Grus antigone NP
(Sarus crane)
Corvus macrorynchos 100 (3/3) 25 (3/12)
(large-billed crow),
n = 1
Gracula religiosa 0 (0/3)
(hill mynah)
Acridotheres tristis 0 (0/4)
(common mynah)
Acridotheres javanicus 0 (0/1)
(white-vented mynah)
Stumus contra (Asian 0 (0/1)
pied starling)
Anhinga melanogaster NP
(oriental darter)
Pelecanus philippensis NP
(spot-billed pelican)
Psittacula eupatria 50 (1/2) 0 (1/146)
(Alexandrine parakeet),
n = 1 ([dagger])
Psittacula roseate 0 (0/20)
(blossom-headed
parakeet)
Psittacula alexandri 0 (0/20)
(red-breasted parakeet)
Psittacula finschii 0 (0/104)
(grey-headed parakeet)
Bubo nipalensis 100 (1/1) 92 (12/13)
(spot-bellied
eagle owl),
n = 1
Ketupa ketupu 100 (3/3)
(buffy fish owl)
Ketupa zeylonensis 86 (6/7)
(brown fish owl)
Strix seloputo 100 (2/2)
(spotted wood owl)
Tyto alba (barn owl) NP
8 sampled 18.3% (44/240)
* S1, aviary section in which cumulative mortality rate could not
be estimated because exact bird population was not known and birds
were difficult to observe; S2, aviary section in which captive bird
population was exactly known and number of dead birds was precisely
recorded; NP, species not present in S1 or S2.
([dagger]) Only sample that was negative for highly pathogenic avian
influenza virus (H5N1); all other birds sampled were positive.
Table 2. Morbidity rates for wild cats during outbreak of highly
pathogenic avian influenza virus (H5N1), Phnom Tamao Wildlife Rescue
Centre, Cambodia, December 15, 2003-January 13, 2004
Order Family Species (common name) Cumulative
morbidity rate,
% (sick/at
risk), no.
sampled
Carnivora Felidae Panthera leo (lion) 100 (2/2)
Panthera tigris (tiger) 80 (8/10), n= 1 *
Catopuma temminckii
(Asiatic golden
cat) 100 (2/2), n = 1 *
Panthera pardus
(leopard) 100 (3/3), n = 1 *
Neofelis nebulosa
(clouded leopard) 100 (1/1), n = 1 *
Prionailurus
bengalensis
(leopard cat) 0 (0/16)
Prionailurus
viverrinus
(fishing cat) 0 (0/5)
Total 41 (16/39)
* All serum samples were positive (date of collection: March 4, 2004).
Copyright:
COPYRIGHT 2009 U.S. National Center for Infectious Diseases
