Perustietoja:
ian Yin, ... Hao Wu, in Handbook of Cell Signaling (Second Edition), 2010
Regulation of TRAF Signaling
TRAF signaling is tightly regulated by many mechanisms; in particular, oligomerization and ubiquitination. These regulatory mechanisms ensure that TRAFs are activated upon ligand stimulation and turned off at the appropriate times. Oligomerization appears to be the common theme of TRAF6 activation in all known signaling pathways: oligomerization by TNFRs, by TIR signaling complexes, and by the CBM complex. The C-terminal TRAF domain mediates its trimerization.
Forced dimerization by fused dimerizing domain also activates TRAF6 [47]. A cytosolic TRAF-interacting protein known as TIFA, with a forkhead-associated (FHA) domain, can enhance TRAF6 oligomerization and activation [48].
Several de-ubiquitinating and ubiquitinating enzymes, such as A20 and CYLD, have been shown to provide feedback inhibition of TRAF-mediated NFκB activation. A20 was originally characterized as an early response gene to TNF stimulation [49], and possesses dual ubiquitin editing functions [50]. While the N-terminal domain of A20 is a de-ubiquitinating enzyme (DUB) for Lys63-linked polyubiquitination of signaling mediators such as TRAF6 and RIP, its C-terminal domain is a ubiquitin ligase (E3) for Lys48-linked degradative polyubiquitination of the same substrates [50–54]. CYLD is Lys63-specific de-ubiquitinating enzyme, whose mutations are the underlying causes of familial cylindromatosis, with predisposition to tumors of skin appendages called cylindromas [44–46, 54, 55].
2. lähde 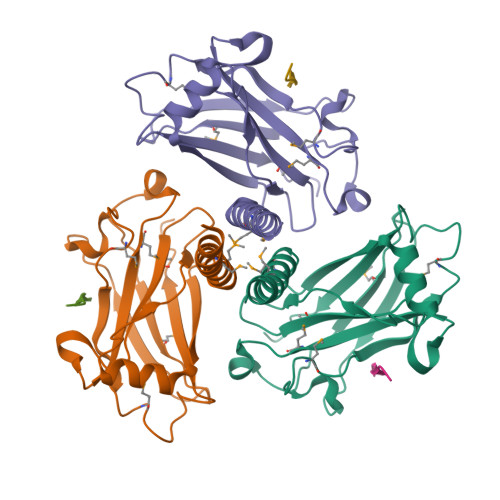
1QSC
CRYSTAL STRUCTURE OF THE TRAF DOMAIN OF TRAF2 IN A COMPLEX WITH A PEPTIDE FROM THE CD40 RECEPTOR
- DOI: 10.2210/pdb1QSC/pdb
- Classification: SIGNALING PROTEIN
- Organism(s): Homo sapiens
- Expression System: Spodoptera frugiperda
- Mutation(s): No
3.lähde
Qian Yin, ... Hao Wu, in Handbook of Cell Signaling (Second Edition), 2010
Regulation of TRAF Signaling
TRAF signaling is tightly regulated by many mechanisms; in particular, oligomerization and ubiquitination. These regulatory mechanisms ensure that TRAFs are activated upon ligand stimulation and turned off at the appropriate times. Oligomerization appears to be the common theme of TRAF6 activation in all known signaling pathways: oligomerization by TNFRs, by TIR signaling complexes, and by the CBM complex. The C-terminal TRAF domain mediates its trimerization. Forced dimerization by fused dimerizing domain also activates TRAF6 [47]. A cytosolic TRAF-interacting protein known as TIFA, with a forkhead-associated (FHA) domain, can enhance TRAF6 oligomerization and activation [48].
Several de-ubiquitinating and ubiquitinating enzymes, such as A20 and CYLD, have been shown to provide feedback inhibition of TRAF-mediated NFκB activation. A20 was originally characterized as an early response gene to TNF stimulation [49], and possesses dual ubiquitin editing functions [50]. While the N-terminal domain of A20 is a de-ubiquitinating enzyme (DUB) for Lys63-linked polyubiquitination of signaling mediators such as TRAF6 and RIP, its C-terminal domain is a ubiquitin ligase (E3) for Lys48-linked degradative polyubiquitination of the same substrates [50–54]. CYLD is Lys63-specific de-ubiquitinating enzyme, whose mutations are the underlying causes of familial cylindromatosis, with predisposition to tumors of skin appendages called cylindromas [44–46, 54, 55].
4. . lähde:
Inga kommentarer:
Skicka en kommentar